NOMIS scientist Jacob Corn has published a study in the journal Cell: “A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation.” A summary follows.
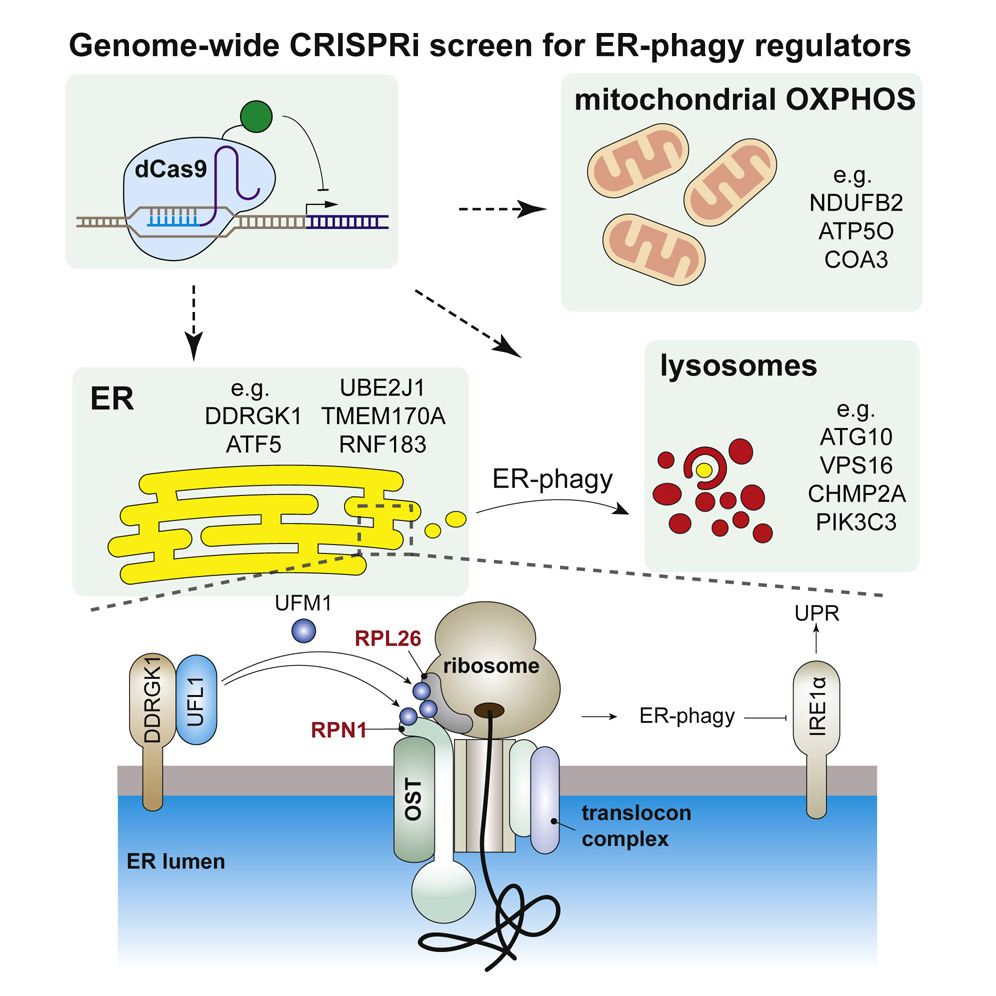
Selective autophagy of organelles is critical for cellular differentiation, homeostasis, and organismal health. Autophagy of the endoplasmic reticulum (ER-phagy) is implicated in human neuropathy but is poorly understood beyond a few autophagosomal receptors and remodelers. By using an ER-phagy reporter and genome-wide CRISPRi screening, we identified 200 high-confidence human ER-phagy factors. Two pathways were unexpectedly required for ER-phagy. First, reduced mitochondrial metabolism represses ER-phagy, which is opposite of general autophagy and is independent of AMPK. Second, ER-localized UFMylation is required for ER-phagy to repress the unfolded protein response via IRE1α. The UFL1 ligase is brought to the ER surface by DDRGK1 to UFMylate RPN1 and RPL26 and preferentially targets ER sheets for degradation, analogous to PINK1-Parkin regulation during mitophagy. Our data provide insight into the cellular logic of ER-phagy, reveal parallels between organelle autophagies, and provide an entry point to the relatively unexplored process of degrading the ER network.
Read the Cell article


