Structure and Function of the Human Ribonucleosome
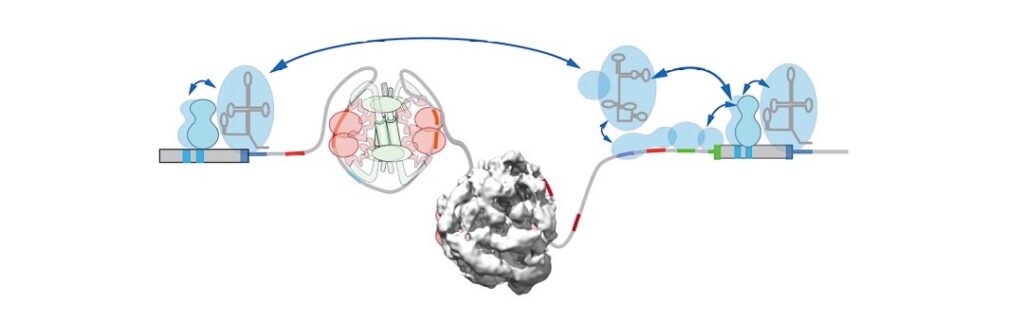
More than 50 years ago, a family of heterogenous nuclear ribonucleoproteins (hnRNPs) was detected and proposed to package and regulate transcript expression. Recently, scientists in the Allain, Blencowe, Mühlemann and […]
Mapping moral language on US presidential primary campaigns reveals rhetorical networks of political division and unity
During political campaigns, candidates use rhetoric to advance competing visions and assessments of their country. Research reveals that the moral language used in this rhetoric can significantly influence citizens’ political attitudes and behaviors; however, the moral language actually used in the rhetoric of elites during political campaigns remains understudied. Using a data set of every tweet (N = 139, 412) published by 39 US presidential candidates during the 2016 and 2020 primary elections, we extracted moral language and constructed network models illustrating how candidates’ rhetoric is semantically connected. These network models yielded two key discoveries. First, we find that party affiliation clusters can be reconstructed solely based on the moral words used in candidates’ rhetoric. Within each party, popular moral values are expressed in highly similar ways, with Democrats emphasizing careful and just treatment of individuals and Republicans emphasizing in-group loyalty and respect for social hierarchies. Second, we illustrate the ways in which outsider candidates like Donald Trump can separate themselves during primaries by using moral rhetoric that differs from their parties’ common language. Our findings demonstrate the functional use of strategic moral rhetoric in a campaign context and show that unique methods of text network analysis are broadly applicable to the study of campaigns and social movements. © The Author(s) 2023. Published by Oxford University Press on behalf of National Academy of Sciences. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted reuse, distribution, and reproduction in any medium, provided the original work is properly cited.
Whole-mouse clearing and imaging at the cellular level with vDISCO
Homeostatic and pathological phenomena often affect multiple organs across the whole organism. Tissue clearing methods, together with recent advances in microscopy, have made holistic examinations of biological samples feasible. Here, we report the detailed protocol for nanobody(VHH)-boosted 3D imaging of solvent-cleared organs (vDISCO), a pressure-driven, nanobody-based whole-body immunolabeling and clearing method that renders whole mice transparent in 3 weeks, consistently enhancing the signal of fluorescent proteins, stabilizing them for years. This allows the reliable detection and quantification of fluorescent signal in intact rodents enabling the analysis of an entire body at cellular resolution. Here, we show the high versatility of vDISCO applied to boost the fluorescence signal of genetically expressed reporters and clear multiple dissected organs and tissues, as well as how to image processed samples using multiple fluorescence microscopy systems. The entire protocol is accessible to laboratories with limited expertise in tissue clearing. In addition to its applications in obtaining a whole-mouse neuronal projection map, detecting single-cell metastases in whole mice and identifying previously undescribed anatomical structures, we further show the visualization of the entire mouse lymphatic system, the application for virus tracing and the visualization of all pericytes in the brain. Taken together, our vDISCO pipeline allows systematic and comprehensive studies of cellular phenomena and connectivity in whole bodies. © 2023, Springer Nature Limited.
Fueling next-generation genome editing with DNA repair
Genome editing technologies generate targeted DNA lesions and rely on cellular DNA repair pathways for resolution. Understanding the DNA repair mechanisms responsible for resolving the specific damage caused by gene editing tools can significantly advance their optimization and facilitate their broader application in research and therapeutic contexts. Here we explore the cellular processes involved in repairing base and prime editor-generated DNA lesions and strategies to leverage and manipulate DNA repair pathways for desired genomic changes. © 2023 The Author(s)
Stochastic motion and transcriptional dynamics of pairs of distal DNA loci on a compacted chromosome
Chromosomes in the eukaryotic nucleus are highly compacted. However, for many functional processes, including transcription initiation, the pairwise motion of distal chromosomal elements such as enhancers and promoters is essential and necessitates dynamic fluidity. Here, we used a live-imaging assay to simultaneously measure the positions of pairs of enhancers and promoters and their transcriptional output while systematically varying the genomic separation between these two DNA loci. Our analysis reveals the coexistence of a compact globular organization and fast subdiffusive dynamics. These combined features cause an anomalous scaling of polymer relaxation times with genomic separation leading to long-ranged correlations. Thus, encounter times of DNA loci are much less dependent on genomic distance than predicted by existing polymer models, with potential consequences for eukaryotic gene expression. © 2023 American Association for the Advancement of Science. All rights reserved.
Johannes Fink, IST Austria scientists demonstrate quantum radar prototype
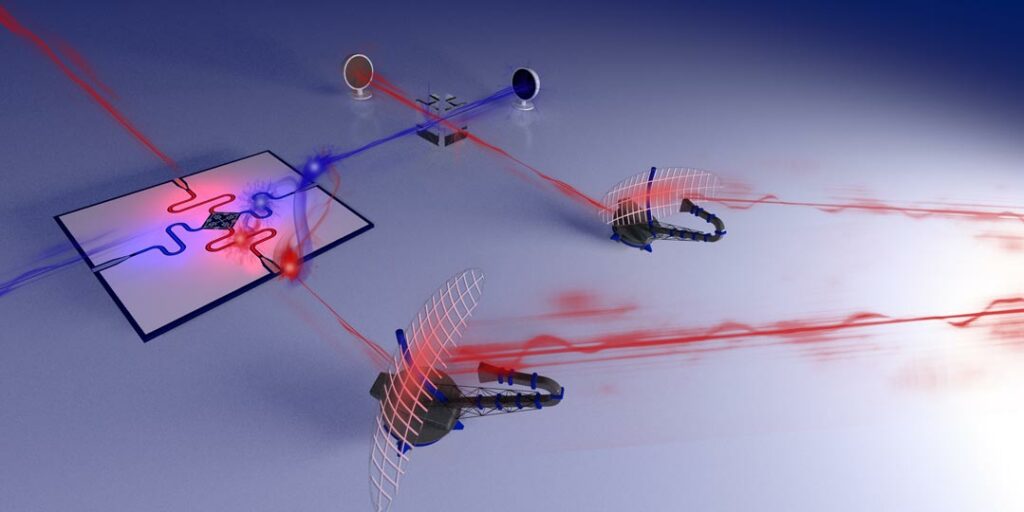
New detection technique based on quantum technology developed at IST Austria – Study published in Science Advances Physicists at the Institute of Science and Technology Austria (IST Austria) have invented […]
NOMIS & Science Young Explorer Award celebrates groundbreaking research by emerging scientists

The 2024 NOMIS & Science Young Explorer Award was presented to three exceptional young researchers — Jasmine Kwasa, Jason Griffin and T. Christina Zhao — for their bold, cross-disciplinary approaches at a […]
Quantum processors: Influencing the trace of “missing electrons” in spin qubits

Amid the race to develop and market practical quantum computers, NOMIS researcher Georgios Katsaros and his group at the Institute of Science and Technology Austria (ISTA) pay particular attention to […]
Elena Conti awarded Jung Prize for Medicine 2025

NOMIS Awardee Elena Conti has been awarded the Jung Prize for Medicine 2025 for her seminal contributions to RNA metabolism. The prize is awarded by the Jung Foundation for Science […]
Janelle Ayres elected to American Academy of Microbiology

NOMIS researcher and Salk Institute Professor Janelle Ayres has been elected to the American Academy of Microbiology’s Fellowship Class of 2024. Fellows of the Academy, an honorific leadership group within […]
2022 NOMIS & Science Young Explorer Award presented at ceremony in Zurich

Recognizing young research talent, the 2022 NOMIS & Science Young Explorer Award was presented to grand prize winner Bill Thompson and finalists Célia Lacaux and Stephen Kissler at a ceremony […]
People’s misperception of “fake faces” for real ones could erode social trust

NOMIS Awardee Manos Tsakiris and colleagues have published their findings in iScience showing that people are more likely to perceive artificially generated faces as real than real ones. The findings […]
NOMIS and Science/AAAS launch Young Explorer Award

The NOMIS Foundation and Science/AAAS have launched a new award for young talent. Through the NOMIS & Science Young Explorer Award, the editors of Science and NOMIS wish to recognize bold young researchers with an […]
Susan Kaech named 2020 AAAS Fellow

Susan Kaech, NOMIS Foundation Chair and director of the NOMIS Center for Immunobiology and Microbial Pathogenesis at the Salk Institute, has been named a 2020 Fellow of the American Association […]
Three NOMIS Awardees recognized as 2025 Clarivate Citation Laureates

NOMIS Awardees Andrea Ablasser, Anthony Hyman and David Autor have been named 2025 Clarivate Citation Laureates, honored for their Nobel-class research and pioneering contributions that have shaped science, society and […]
Wolfgang Busch

Wolfgang Busch is a 2025 NOMIS Awardee, Hess Chair in Plant Science, professor and director of the Plant Molecular and Cellular Biology Laboratory at the Salk Institute for Biological Studies […]
Were large soda lakes the cradle of life?

Scientists have long puzzled over how life could have emerged when phosphorus, a key ingredient for DNA, RNA, and cellular energy, was so scarce on early Earth. New research by […]
How antidepressants can protect against infections and sepsis
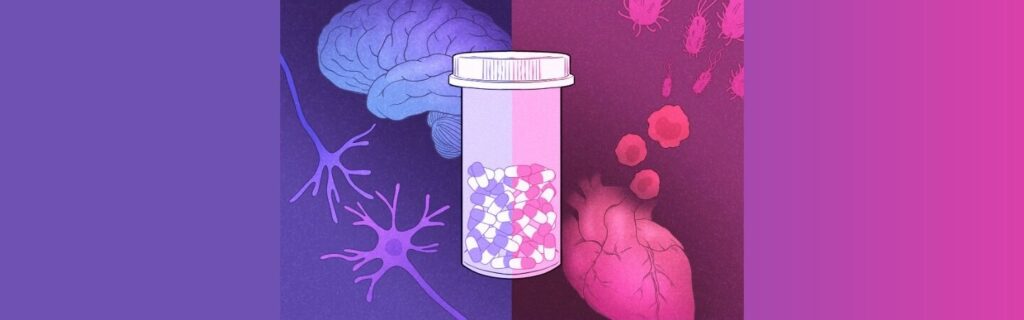
NOMIS researcher Janelle Ayres, together with a team of scientists at the Salk Institute, have discovered how the SSRI Prozac regulates the immune response and prevents sepsis in mice, demonstrating […]
Svante Pääbo featured in newest NOMIS Insight film
NOMIS Awardee and Nobel laureate Svante Pääbo has been featured in the latest NOMIS Insight film, which details the journey of his recently concluded research project, A Cell and Molecular Approach […]
Celebrating the 2023 NOMIS & Science Young Explorer Award winners

Recognizing young research talent, the 2023 NOMIS & Science Young Explorer Award was presented to grand prize winner Michael Skinnider and finalists Isabella Bower and George Goshua at a ceremony […]
2023 NOMIS & Science Young Explorer Award winners announced

Science/AAAS and the NOMIS Foundation have announced the winners of the 2023 NOMIS & Science Young Explorer Award, which recognizes bold early-career researchers who ask fundamental questions at the intersection […]

